Do you have unusual, difficult or valuable samples that cannot or should not be processed in a standard workflow? StarSEQ and its qualified staff offer a service that goes beyond standard workflows. We discuss with you the optimal strategy for your samples and adapt our workflows to your samples.
Life Science/Next Generation Sequencing/
Microbiome Analysis
Amplicon Metagenomic Sequencing and Barcoding
StarSEQ has been the industry leader in this service for over 12 years now. We invite you to benefit from our experience with below special offers.
Incredible 16S and ITS 1 Microbiome Analysis Promotion
Flexible Voucher Option: Save your Funds and Budgets!
Buy now and perform this or any other project within 12 months.
“All in One Microbiome” Packages (available for 16S and ITS 1): High Throughput Amplicon Generation and Sequencing using Special Single Step Amplicon Generation with reduced bias!
Package 384: 384 samples as batch only 23,77 € per sample and region
Package A: 288 samples as batch only 26,52 € per sample and region
Package B: 192 samples as batch only 31,54 € per sample and region
Package C: 96 samples as batch only 47,08 € per sample and region
Package D: 48 samples as batch only 76,17 € per sample and region
Price examples, all other sample amounts are possible. The price is a combination of a fixed price for the run and a variable price for the libraries depending on the number of samples.
No shared runs, customer samples are sequenced exclusively on one or more MiSeq runs!
2-Step amplicon generation and barcoding available for: custom primer, ITS 1/2, various ITS 2, various 18S, Cytb, LSU, SSU, CO1, mcrA, and 16S V1-V2.

Microbiome sequencing has become an indispensable tool in understanding the complex communities of microorganisms inhabiting various environments, including soil, water, the human body, and industrial settings. Three primary molecular markers employed in microbiome studies are 16S ribosomal RNA (16S rRNA), Internal Transcribed Spacer (ITS), and 18S ribosomal RNA (18S rRNA). Each marker targets specific domains of life—bacteria and archaea (16S rRNA), fungi (ITS), and eukaryotes (18S rRNA)—allowing researchers to profile diverse microbial communities with precision.
Chose one option for amplicon sequencing or combine bacterial and fungal microbiome analysis
(1 amplicon = 1 sample):
Single Step Bacterial and Archaea microbiome analysis:
16S, V4 region, Primer combination: 515F (5′-GTGYCAGCMGCCGCGGTAA-3′, recommended) optional (5′-GTGNCAGCMGCCGCGGTAA-3′, old version) – 806bR (5′-GGACTACNVGGGTWTCTAAT-3′)* or
16S, V4-V5 region, Primer combination: 515F (5′-GTGYCAGCMGCCGCGGTAA-3′) – 926R (5′-CCGYCAATTYMTTRAGTTT-3′)* (recommemded) or
16S, V4-V5 region, Primer combination: 515F (5′-GTGNCAGCMGCCGCGGTA-3′) – 909R (=924R) (5′-CCCCGYCAATTCMTTTRAGT-3′)* (old version) or
16S, V3-V4 region, Primer combination: 341f (5′-CCTACGGGNGGCWGCAG-3′) – 806R (=786R) (5′-GACTACNVGGGTWTCTAATCC-3′)**** (recommended) or
16S, V3-V4 region, Primer combination: 341f (5′-CCTACGGGNBGCASCAG-3′) – 806bR (5′-GGACTACNVGGGTWTCTAAT-3′)**** (old version)
or Single Step Fungal (18S) microbiome analysis:
ITS, ITS 1 region, Primer combination: ITS1F (5-CTTGGTCATTTAGAGGAAGTAA-3′) – ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′)** (Note: ITS1F primer (Gardes and Bruns, 1993) is 38 bp upstream of ITS1 from White et al., 1990)
! Please note that for 16S and other low diversity regions the quality of read 2 is lower than read 1. This is not an sequencing or quality error!
or 2-Step Bacterial, Archaea and Methanogens microbiome analysis:
16S, V1-V2 region, Primer combination 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) – 338R (5′-TGCTGCCTCCCGTAGGAGT-3′)
16S, V4 region, Primer combination 528F (5′-GCGGTAATTCCAGCTCCAA-3′) – 706R (5′-AATCCRAGAATTTCACCTCT-3′)
mcrA, Primer combination mlas-For (5′-GGTGGTGTMGGDTTCACMCARTA-3′) – mcrA-Rev (5′-CGTTCATBGCGTAGTTVGGRTAGT-3′)‡
or 2-Step Fungal (18S) microbiome analysis:
ITS, ITS 1/2 region, Primer combination: ITS-u2 (5′-GAAYCATCGARTCTTTGAACGC-3′) – ITS-p4 (5′-CCGCTTAKTGATATGCTTAAA-3′)
ITS, ITS 2 region, Primer combination: ITS3F (5′-GCATCGATGAAGAACGCAGC-3′) – ITS4R (5′-TCCTCCGCTTATTGATATGC-3′)**
ITS, ITS 2 region, Primer combination: 5.8S-Fun (5′-AACTTTYRRCAAYGGATCWCT-3′) – ITS4-Fun (5′-AGCCTCCGCTTATTGATATGCTTAART-5′)‡‡‡‡
ITS, ITS 2 region, Primer combination: fITS7 (5′-GAGACAGGTGARTCATCGAATCTTTG-3′)‡‡‡‡‡ – ITS4R (5′-TCCTCCGCTTATTGATATGC-3′)**
SSU region, SSUfungiF (5′-TGGAGGGCAAGTCTGGTG-3′) – SSUFungiR (5′-TCGGCATAGTTTATGGTTAAG-3′)
LSU region, LSU Fungi LR0R_f (5′-ACCCGCTGAACTTAAGC-3′) – LSU Fungi JH-LSU-369rc_r (5′-CTTCCCTTTCAACAATTTCAC-3′)
AMF (arbuscular mycorrhiza fungi), NS31-For (5′-TTGGAGGGCAAGTCTGGTGCC-3′)‡‡ – AML2-Rev (5′-GAACCCAAACACTTTGGTTTCC-3′)‡‡‡
or 2-Step Eukaryotes (microbial) analysis:
V4–V5 region, Primer combination: 574-f (5′-CGGTAAYTCCAGCTCYV-3′) – 1132R (5′-CCGTCAATTHCTTYAART-3′)‡‡‡‡‡‡
V8-V9 region, Primer combination: 18S-1422f (5′-ataacaggtctgtgatgccct-3′) – 18S_1797R (5′-ccttcygcaggttcacctac-3′)*****
V9 region, Primer combination: Illumina_Euk_1391f (5′-GTACACACCGCCCGTC-3′) – Illumina_EukBr_1510r (5′-TGATCCTTCTGCAGGTTCACCTAC-3′), blocking primer optional***; V9 region samples could not be combined with ITS, V8-V9 or 16S samples and need to be sequenced in a seperate MiSeq run.
or 2-Step Barcoding:
cytochrome b, cytb1 (5′-ccatccaacatctcagcatgatgaaa-3′) – cytb2 (5′-gcccctcagaatgatatttgtcctca-3′)
cytochrome oxidase I, CO I E (5′-ccagagattagagggaatcagtg-3′) – CO 1 F (5′-cctgcaggaggaggagaycc-3′)
or 2-Step Custom Amplicons:
Customer specific assays, design of custom primers, request your special quotation: info@starseq.com
Send us your gDNA samples and make use of our “All in One” Service:
• Quality control
• Single step amplicon generation with reduced bias
• Double indexing, quality check, quantification, normalization and pooling of amplicons
• Illumina MiSeq sequencing package A-C (16S + ITS): exclusive 2 x 300 nt paired-end sequencing with V3 chemistry
• Illumina MiSeq sequencing package A-C (18S): exclusive 2 x 250 nt paired-end sequencing with V2 chemistry full flow cell
• Illumina MiSeq sequencing package D (16S + ITS + 18S): exclusive 2 x 250 nt paired-end sequencing with V2 chemistry nano flow cell
• Output for package A-C (16S + ITS): 20-30 M reads (including up to 25 % PhiX to balance the composition of bases)*
• Output for package A-C (18S): 16-20 M reads (including up to 25 % PhiX to balance the composition of bases)*
• Output for package D (16S + ITS + 18S): 750 K reads (including up to 25 % PhiX to balance the composition of bases)*
• De-multiplexing of reads
• Data delivery via FTP server
*Please note that for 16S and other low diversity samples the output is lower than Illumina specifies for standard chemistry V2 and V3 runs.
Applications:
• Environmental Metagenomics (soil, water, air, biofilms, complex organic communities): Understanding nutrient cycling, soil fertility, and the impact of agricultural practices on microbial communities, studying microbial dynamics in freshwater and marine environments, including the effects of pollution and climate change
• Human or Animal Microbiome (skin, stool, gut, blood, swab, tissue, tumour): Exploring interactions among microorganisms and their roles in health.
• Monitoring of Animal Health (e.g. aternatively or subblementerily to FELASA-Test)
• Sterility Monitoring
• Detection of Contamination
• Monitoring of Biogas Plant and Bioreactors: Optimizing fermentation processes by monitoring microbial consortia in bioreactors
• Biosafety Monitoring
• Food Quality: Ensuring product quality and safety by analyzing microbial populations in production environments.
• Clinical Samples
Sequencing of your ready to load 16S/ITS/18S libraries:
• Illumina MiSeq sequencing: 2 x 300 nt paired-end sequencing with V3 chemistry or 2 x 250 nt paired-end sequencing with V2 chemistry
• V3 chemistry output is 20-30 M reads (including up to 25 % PhiX to balance the composition of bases)
• V2 chemistry output 16-20 M reads (including up to 25 % PhiX to balance the composition of bases)
• De-multiplexing of reads
• Data delivery via FTP server
Only 3300 €
Primer sequence references:
*Apprill A, McNally S, Parsons R, Weber L. 2015. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol 75:129–137. Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18: 1403–1414.
*Parada, A. E., Needham, D. M., & Fuhrman, J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environmental Microbiology, 18(5), 1403–1414. https://doi.org/10.1111/1462-2920.13023
*Walters, W., Hyde, E. R., Berg-Lyons, D., Ackermann, G., Humphrey, G., Parada, A., … Knight, R. (2016). Improved Bacterial 16S rRNA Gene (V4 and V4-5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems, 1(1), e00009-15. https://doi.org/10.1128/mSystems.00009-15
**White, T. J., T. Bruns, S. Lee, and J. W. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. Pp. 315-322 In: PCR Protocols: A Guide to Methods and Applications. Academic Press, New York, NY. Gardes, M., and T. D. Bruns. 1993. ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Mol. Ecol. 2: 113-118.
**Gardes, M., & Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes ‐ application to the identification of mycorrhizae and rusts. Molecular Ecology, 2(2), 113–118.
***Amaral-Zettler, L. A., McCliment, E. A., Ducklow, H. W., & Huse, S. M. (2009). A Method for Studying Protistan Diversity Using Massively Parallel Sequencing of V9 Hypervariable Regions of Small-Subunit Ribosomal RNA Genes. PLOS ONE, 4(7), e6372. Retrieved from https://doi.org/10.1371/journal.pone.0006372
**** Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., et al. . (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, 1–11. 10.1093/nar/gks808
‡ Steinberg LM, Regan JM 2008. Phylogenetic Comparison of the Methanogenic Communities from an Acidic, Oligotrophic Fen and an Anaerobic Digester Treating Municipal Wastewater Sludge. Appl Environ Microbiol 74: https://doi.org/10.1128/AEM.00553-08
1992. Specific amplification of 18S fungal ribosomal genes from vesicular-arbuscular endomycorrhizal fungi colonizing roots. Appl Environ Microbiol 58: https://doi.org/10.1128/aem.58.1.291-295.1992
‡‡‡ Jaikoo Lee, Sangsun Lee, J. Peter W. Young, Improved PCR primers for the detection and identification of arbuscular mycorrhizal fungi, FEMS Microbiology Ecology, Volume 65, Issue 2, August 2008, Pages 339–349, https://doi.org/10.1111/j.1574-6941.2008.00531.x
‡‡‡‡ 2016. Accurate Estimation of Fungal Diversity and Abundance through Improved Lineage-Specific Primers Optimized for Illumina Amplicon Sequencing. Appl Environ Microbiol 82:. https://doi.org/10.1128/AEM.02576-16
‡‡‡‡‡ Katarina Ihrmark, Inga T.M. Bödeker, Karelyn Cruz-Martinez, Hanna Friberg, Ariana Kubartova, Jessica Schenck, Ylva Strid, Jan Stenlid, Mikael Brandström-Durling, Karina E. Clemmensen, Björn D. Lindahl, New primers to amplify the fungal ITS2 region – evaluation by 454-sequencing of artificial and natural communities, FEMS Microbiology Ecology, Volume 82, Issue 3, December 2012, Pages 666–677, https://doi.org/10.1111/j.1574-6941.2012.01437.x
‡‡‡‡‡‡ Systematic Design of 18S rRNA Gene Primers for Determining Eukaryotic Diversity in Microbial Consortia Hugerth LW, Muller EEL, Hu YOO, Lebrun LAM, Roume H, et al. (2014) Systematic Design of 18S rRNA Gene Primers for Determining Eukaryotic Diversity in Microbial Consortia. PLOS ONE 9(4): e95567. https://doi.org/10.1371/journal.pone.0095567
see also https://support.illumina.com/downloads/16s_metagenomic_sequencing_library_preparation.html
Simply order by e-mail: microbiome@starseq.com
You have no time/staff for routine preparation of gDNA from your collected samples? As an additional service we offer also the purification of gDNA from various starting material. Please contact us if you are interested in further service options or other gene regions and we will find YOUR optimal solution!
info@starseq.com
Advanced and complex bioinformatic analysis of 16S/18S/ITS sequences (QIIME 2)
The bioinformatics analysis pipeline consists of the following steps including quality control, pre-processing of reads, taxonomy assignments and visualization, calculation of alpha and beta-diversity (optional) metrics.
- Raw reads are de-multiplexed and quality checked by FastQC¹.
- Primers are trimmed using the tool cutadapt² if nescessary.
- Paired-end reads are joined by the tool PEAR or VSEARCH³.
- Low-quality reads are removed.
- Reads are corrected, chimeras are removed and Amplicon Sequence Variants (ASVs) are obtained by the deblur workflow4.
- 16S/18S: A multiple sequence alignment and a phylogenetic tree is generated.
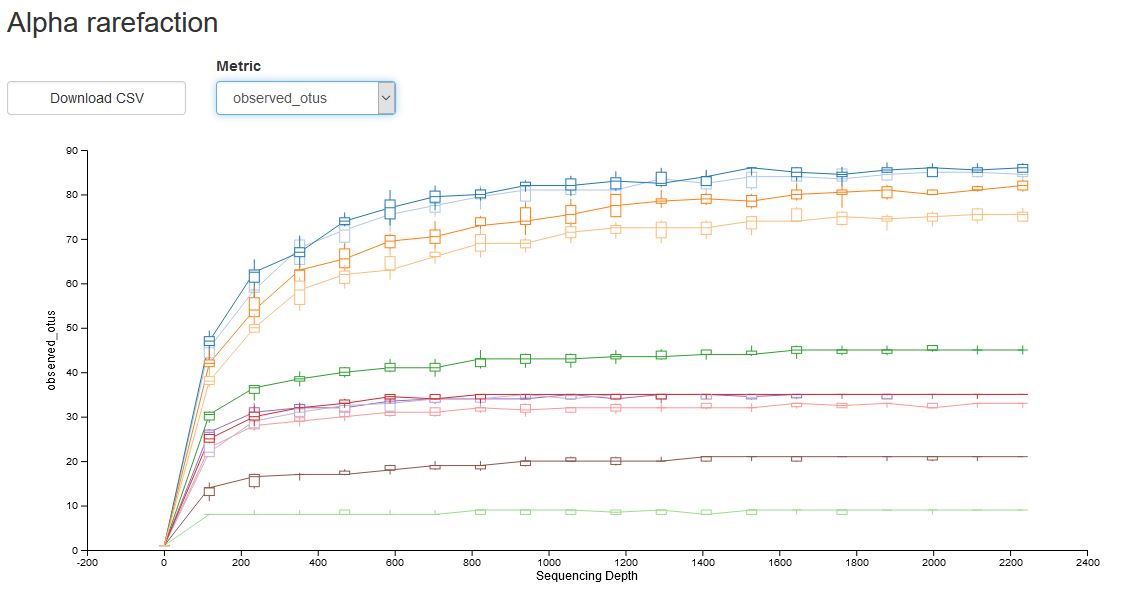
- Alpha-diversity rarefaction curves are generated for categories and each individual sample.
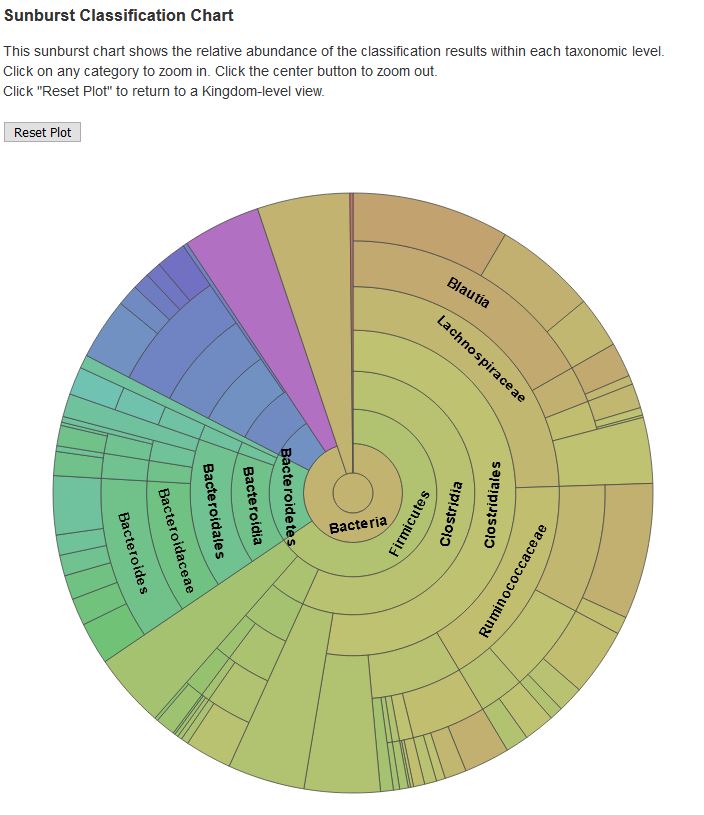
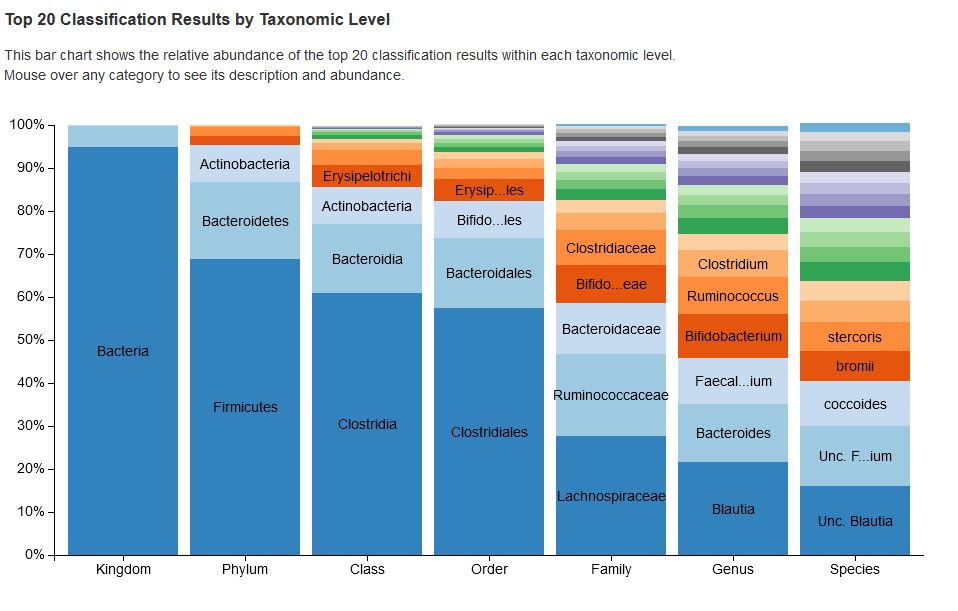
- Taxonomy is assigned to ASVs using a Naive-Bayes approach of the scikit-learn Python library7and the SILVA 13813 (16S/18S) or UNITE (ITS) database. Interactive stacked bar-charts of the taxonomic abundances of each category and each sample are generated.
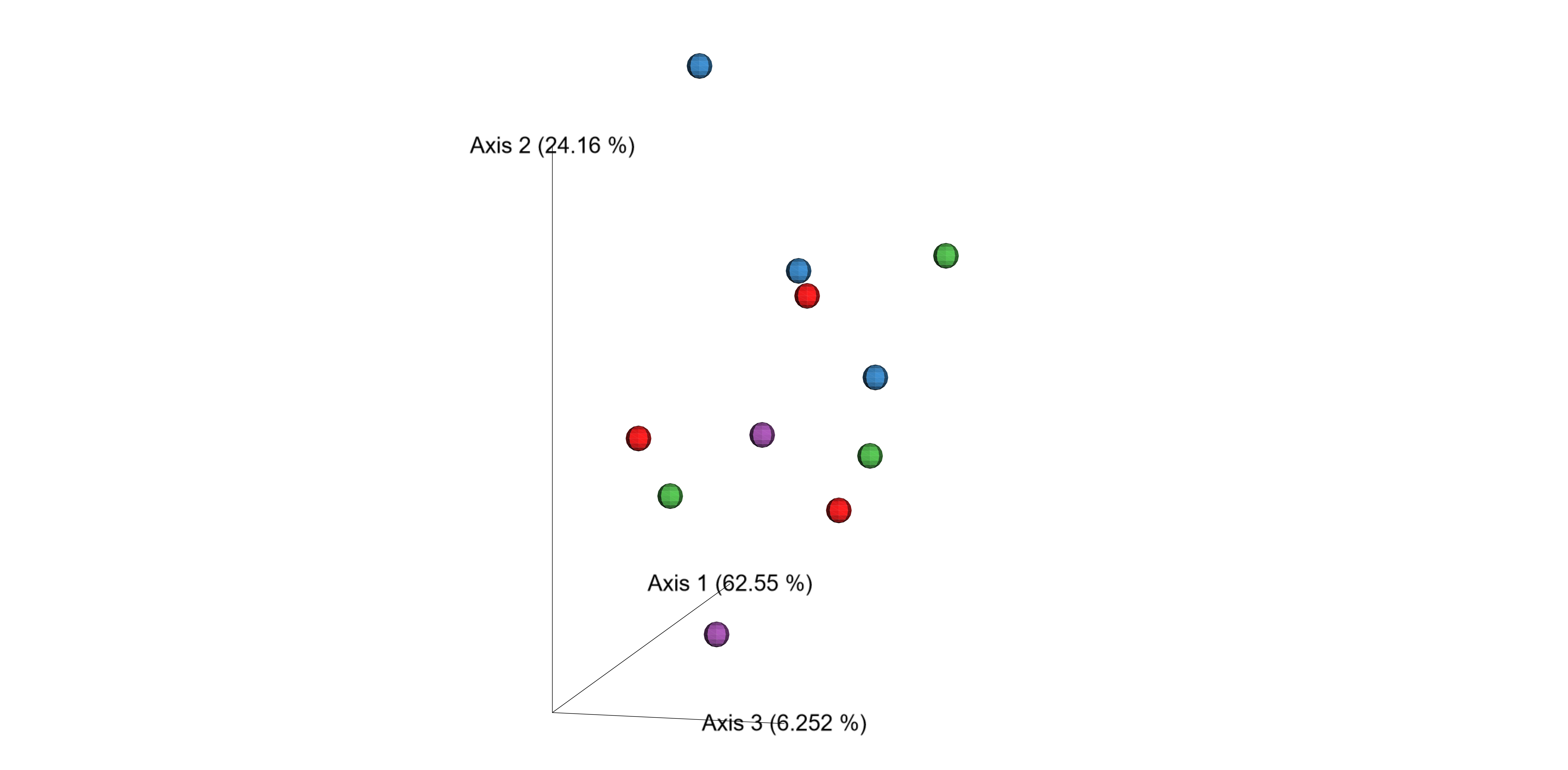
- Alpha and beta diversity metrics are calculated after normalization by rarefaction. Alpha-diversity Shannon metric boxplot are generated comparing different categories.
Output files:
- Dna-sequences.fasta: dna sequences in fasta format including representative sequences of amplicon sequence variants (ASVs).
- Feature-table.biom: biom tables (Biological Observation Matrix: matrix of counts of observations (ASVs) per sample).
- Classification.qza: artifact of Taxonomy.tsv (taxonomic classification of reads).
- Classification.qzv: visualization of taxonomic read classification.
- Shannon’s diversity index (a quantitative measure of community richness).
- Observed OTUs (a qualitative measure of community richness).
- Faith’s Phylogenetic Diversity (a qualitative measure of community richness that incorporates phylogenetic relationships between the features).
- Evenness (or Pielou’s Evenness; a measure of community evenness).
- Jaccard distance (a qualitative measure of community dissimilarity).
- Bray-Curtis distance (a quantitative measure of community dissimilarity).
- Unweighted UniFrac distance (a qualitative measure of community dissimilarity that incorporates phylogenetic relationships between the features).
- Weighted UniFrac distance (a quantitative measure of community dissimilarity that incorporates phylogenetic relationships between the features).
Notes:
Taxonimical assignment is performed down to species level if possible. Due to reasons like sample quality, genus and family diversity and database composition the assignment might only be possible down to genus or family.
Since the ITS region shows more variation than 16S/18S a reliable multiple sequence alignment is not possible11. Therefore the alignment and the tree is not generated for this region and analyses based on diversity matrices (e. g. UniFrac12) leading to erroneous results are not carried out.
- Andrews S. (2010) FastQC: a quality-control tool for high-throughput sequence data. Babraham Institute, Cambridge, United Kingdom.
- Martin, M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. journal, 17(1), pp. 10-12.
- Rognes T., Flouri T., Nichols B., Quince C., Mahé F. (2016) VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
- Amir A., McDonald D., Navas-Molina J.A., Kopylova E., Morton J.T., Zech Xu Z., Kightley E.P., Thompson L.R., Hyde E.R., Gonzalez A., Knight R. (2017) Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2:e00191-16. https://doi.org/10.1128/mSystems.00191-16.
- Katoh, K., Misawa, K., Kuma, K., & Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic acids research, 30(14), 3059–3066.
- Price, M. N., Dehal, P. S., & Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Molecular biology and evolution, 26(7), 1641–1650. doi:10.1093/molbev/msp077
- Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., … Gregory Caporaso, J. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome, 6(1), 90. doi:10.1186/s40168-018-0470-z
- Nilsson, R. H., Larsson, K. H., Taylor, A., Bengtsson-Palme, J., Jeppesen, T. S., Schigel, D., … Abarenkov, K. (2018). The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic acids research, 47(D1), D259–D264. doi:10.1093/nar/gky1022
- Mandal, S., Van Treuren, W., White, R. A., Eggesbø, M., Knight, R., & Peddada, S. D. (2015). Analysis of composition of microbiomes: a novel method for studying microbial composition. Microbial ecology in health and disease, 26, 27663. doi:10.3402/mehd.v26.27663
- Halwachs B., Madhusudhan N., Krause R., Nilsson R.H., Moissl-Eichinger C., Högenauer C., Thallinger G.G., Gorkiewicz G. Critical Issues in Mycobiota Analysis. Microbiol. 2017;8:180.
- Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. 2011 UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011 Feb; 5(2):169-72.
- Chen, J.; Bittinger, K.; Charlson, E. S.; Hoffmann, C.; Lewis, J.; Wu, G. D.; Collman, R. G.; Bushman, F. D.; Li, H. (2012). “Associating microbiome composition with environmental covariates using generalized UniFrac distances”. Bioinformatics. 28 (16): 2106–2113. doi:10.1093/bioinformatics/bts342. PMC 3413390. PMID 22711789
- SILVA terms of use/license information: https://www.arb-silva.de/silva-license-information/. Silva citing:
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl. Acids Res. 41 (D1): D590-D596. Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glöckner FO (2014) The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks.
Nucl. Acids Res. 41 (D1): D590-D596. Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glöckner FO (2014) The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks.  Nucl. Acids Res. 42:D643-D648 Glöckner FO, Yilmaz P, Quast C, Gerken J, Beccati A, Ciuprina A, Bruns G, Yarza P, Peplies J, Westram R, Ludwig W (2017) 25 years of serving the community with ribosomal RNA gene reference databases and tools.
Nucl. Acids Res. 42:D643-D648 Glöckner FO, Yilmaz P, Quast C, Gerken J, Beccati A, Ciuprina A, Bruns G, Yarza P, Peplies J, Westram R, Ludwig W (2017) 25 years of serving the community with ribosomal RNA gene reference databases and tools.  J. Biotechnol.
J. Biotechnol. - QIIME 2: Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, and Caporaso JG. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology 37: 852–857. https://doi.org/10.1038/s41587-019-0209-9
Basic bioinformatic analysis of 16S/ITS sequences (Illumina-App)
The bioinformatics analysis performs taxonomic classification of 16S/ITS rRNA targeted amplicon reads using the RefSeq RDP 16S v3 taxonomic database and the UNITE Fungal ITS Database v7.2
The Illumina-app provides interactive visualizations and raw classification output for per-sample and aggregate analyses.
Wang Q. et al (http://dx.doi.org/10.1128%2FAEM.00062-07)
Ali Alishum. (2019). DADA2 formatted 16S rRNA gene sequences for both bacteria & archaea (Version Version 2)
UNITE Community.(https://doi.org/10.15156/BIO/587475)
Microbial DNA preparation service from various starting material:
StarSEQ has long term and advanced knowledge in extracting DNA from a wide range of starting material. We adjust varied DNA extraction protocols, kits and bead mill grinding to meet the individual demands of any project. We are using the most advanced tissue homogenizer from Bertin (Precellys Evolution and Minilys) for optimal adapted sample preparation.
Our experience covers almost all kind of genetic material:
• All kind of tissues and blood
• Swabs from skin and other surfaces
• Fecal samples
• Saliva samples
• Soil and sluge
• Sediments
• Plant tissues, roots, seeds or leafs
• Fungi tissues and spores
• Water and liquids
• Air filter
• Biofilms
• Nutriments
Starting from 12€/sample

Sequencing of your ready to load 16S/ITS/18S libraries:
• Illumina MiSeq sequencing: 2 x 300 nt paired-end sequencing with V3 chemistry
• Output 20-30 M reads (low cluster density including 25 % PhiX to balance the composition of bases)
• De-multiplexing of reads
• Free 16S metagenomics analysis (Illumina App)
• Data delivery via FTP server
Only 3300 €