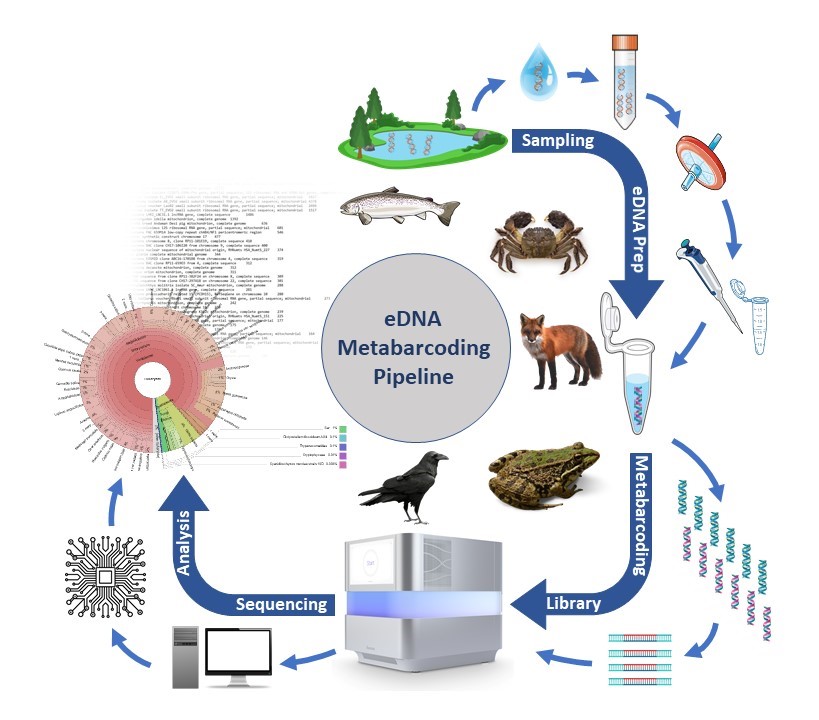
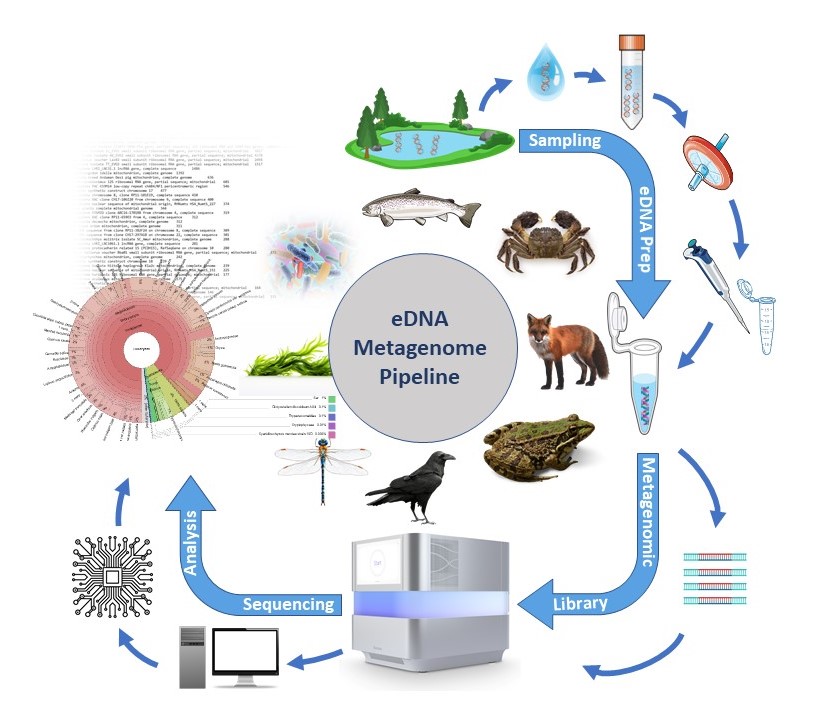
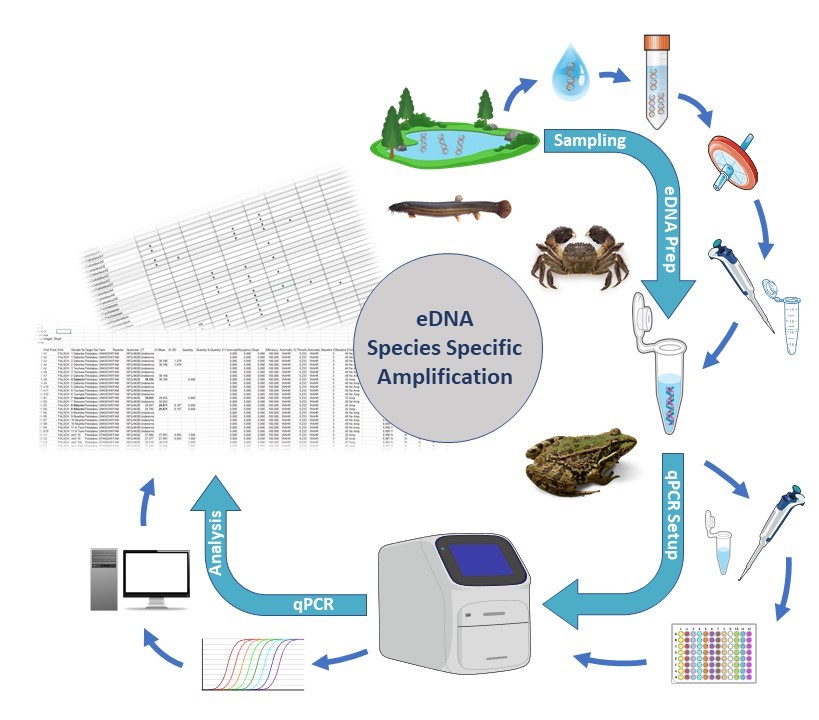
What is environmental DNA?
The term ‘environmental DNA’ (eDNA) refers to the free DNA released by all living organisms into their environment. Monitoring using eDNA in bodies of water is a minimally invasive method, as it does not require the target organisms to be caught or removed from the water. The premise of this method is that every organism releases DNA into its environment through cell abrasion, excreta or secretion of DNA.
To illustrate this point, consider that a fish of 30-75g (for example, a small perch) releases more than 107 copies of DNA per hour into the surrounding water. However, it is important to note that DNA is also subject to permanent degradation by various environmental influences.Indeed, the eDNA half-life was calculated to be around 6 hours, which indicates that more than 90% of eDNA copies will have degraded within 24 hours (Maruyama, 2014). Furthermore, DNA fragments measuring up to 300-400bp remain identifiable within the water for a period of only 1-2 weeks following the species’ removal (Dejean, 2011).The quantity of eDNA in the water undergoes a rapid decrease, meaning that a species can only be detected in the water if it is currently present or has just been present.
The result of an eDNA analysis is a snapshot. While DNA in nature can endure for extended periods, the detection of free-floating DNA fragments remains contingent on variables such as flow velocity, duration, and the true number of individuals, which can also be influenced by seasonal factors.