SARS-CoV-2 Genome Sequencing: Uncover the Virus Variants – Implementation of sequencing workflow in Just One Week
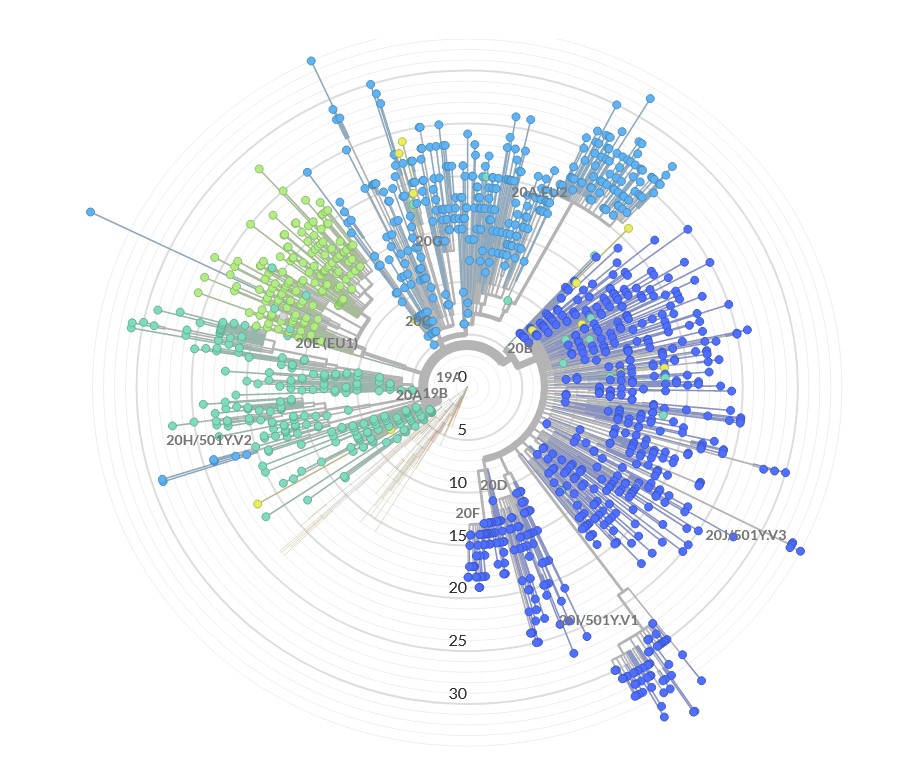
Together with with the Johannes Gutenberg University Mainz (Institute of Organismic and Molecular Evolution, Molecular Genetics & Genome Analysis) and the University Medical Center (Institute for Virology and Research Center for Immunotherapy (FZI), the Institute for Humangenetics, the Department of Hygiene and Infection Prevention – Hospital Hygiene -, the Medical Management Department and the Transfusion Unit & Test Center) StarSEQ founded the SARS-CoV-2 Sequencing Consortium Mainz. The goal of this high-level scientific cooperation is the massive parallel sequencing of SARS-CoV-2 genomes in rhineland-palatinate and the rhine-main area to uncover genetic changes (mutations) of the Covid-19 virus for regional SARS-CoV-2 variant surveillance. We have uploaded the sequencing and bioinformatic analyis data to the RKI (Robert Koch Institut) and the GISAID Initiative. Read the press release here (German). We are also participating in the project “Inclusion of molecular methods for fine characterization and direct detection of polioviruses from wastewater samples (PIA II)” of the National Reference Center for Poliomyelitis and Enteroviruses at the Robert Koch Institute!
Update: Here you can read the preprint of our newest puplications: